How can the philosophy of science help inform our response to COVID-19? Stephan Guttinger looks at the history and philosophy of ribonucleic acid (RNA), a central but often overlooked molecule in the story of the pandemic.
The history and philosophy of the life sciences (HPLS) offers powerful resources for the analysis of, and response to, the COVID-19 pandemic. Reflections on evolutionary theory, the history of vaccination, or the power and limitations of animal model research (to name just a few examples) can all contribute to a better understanding of the pandemic. However, key elements of the pandemic have to date gained little attention in HPLS. One of these elements is ribonucleic acid (RNA). Given the central role of RNA, developing a history and philosophy of this molecule will provide HPLS scholars with important tools to analyse the unfolding of the pandemic and science’s response to it. This will also enable scholars to build a more detailed understanding of the contemporary life sciences, as RNA is becoming a more central element in biological theory and practice.
RNA and the behaviour of viruses
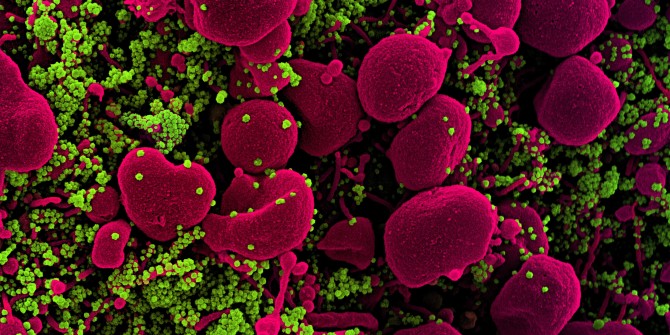
The first and probably most prominent reason why RNA matters for the pandemic can be found in the genomics of SARS-CoV-2: like many other viruses – and unlike plants, animals, or bacteria – the genome of this virus consists of an RNA molecule rather than a DNA molecule. This difference matters for several reasons. First, it matters because RNA, unlike DNA, can form intricate three-dimensional structures on its own, ranging from simple hairpins to more elaborate higher-order arrangements. These structural features affect how RNA genomes are packaged and regulated, which in turn affects how viruses behave and respond to interventions.
To capture the special nature of RNA genomes, virologists now often talk of two layers of information they carry: the sequence layer and the structural layer. To analyse these layers and their functional relevance, researchers develop new techniques, quickly reshaping the conceptual and the methodological landscape of molecular biology.
Another important feature of most RNA viruses is their high mutation rate. This high rate of change at the sequence level means that any viral population will consist of a broad spectrum of variants. This has led virologists to use terms such as “quasispecies” or “mutant cloud” (rather than traditional terms such as “species”) when they describe RNA viruses.
The use of new metaphors such as “mutant cloud” might seem innocuous at first, but it has important consequences for scientific practice and theory. When researchers speak of “clouds” they shift their focus from thinking in terms of distinct and well-defined viral particles to thinking about a more distributed and dynamic entity. Cloud members interact with each other and with the cloud’s environment. This interconnectedness is functionally relevant, as it can affect viral fitness or even features that are usually seen as “intrinsic” properties of RNA viruses, such as their mutation rate. In the traditional picture of viruses, each virus simply has “its” mutation rate, depending on the type of (error-prone) RNA polymerase that it encodes in its genomic sequence. Recent research on the actual mutation rates of RNA viruses (qua mutant clouds), however, has shown that the mutation rate is a more complex feature of a population that is also set to a large degree by cellular processes (such as RNA editing) that interact with the viral cloud.
This highly dynamic and context-dependent nature of RNA viruses also seems to matter in the case of SARS-CoV-2. It has been shown that SARS-CoV-2 is a quasispecies with high sequence diversity and dynamics, even within single patients. At the same time, there are indications that this virus does not mutate as fast as other RNA viruses and that the quasispecies can be surprisingly stable when the virus is replicated in cultured cells. These findings indicate that the dynamics of SARS-CoV-2 are co-determined by the system within which it exists, in line with what a cloud-approach to viruses would predict.
In order to get a better understanding of SARS-CoV-2 and of how scientists study this emerging virus, it will therefore be important to gain a better understanding of concepts such as “viral clouds” and, more generally, of how RNA is changing the conceptual and methodological landscape of contemporary research.
RNA as a key tool to study and fight viruses
It is not just the biology of viruses that makes RNA an important topic for HPLS. A second reason is that RNA has emerged as a powerful research tool that transforms the way in which scientists study biological systems.
Underlying the growing power of RNA as a research tool is again the fact that RNA molecules can fold into three-dimensional structures. This makes RNA a unique structural glue: rather than serving as a simple messenger that connects DNA with the protein translation machinery – an important but limited role that RNA was demoted to for decades in biological theory – RNA now emerges as a structurally complex guide for a range of DNA- or RNA-modifying enzymes, targeting them to specific sites on cellular or viral genomes.
The discovery of such “non-coding” RNAs (so called because they don’t code for protein products) has reshaped how scientists understand the working of cells, the development of organisms, and the dynamics of evolution. These discoveries have also had important methodological implications for how researchers study biological systems, including SARS-CoV-2. Adapting RNA as a tool in the laboratory has allowed researchers to quickly and cheaply “re-programme” enzyme complexes, targeting them to specific loci simply by changing the sequence of the non-coding guide RNA. This principle of programmable enzyme complexes is used for RNA interference (RNAi) and also for novel gene editing tools, such as CRISPR-Cas9.
These tools allow researchers to manipulate the expression of specific genes or to re-write the genetic code of model systems, thus revolutionising how scientists can manipulate their objects of interest.
The idea of using RNA to re-programme biological processes is also used in the development of novel vaccines. Rather than injecting, for instance, an attenuated virus or parts of a virus particle, researchers now inject an RNA that codes for viral proteins, forcing the cell to produce viral elements which then (hopefully) trigger an immune response. This is also one of the leading technologies currently used to quickly produce vaccines against SARS-CoV-2.
The impact that RNA has had on research practice suggests that the dynamics, the power, and the limitations of the life sciences cannot be understood without also taking into account RNA biology. At the same time, ever since RNA has emerged as a key player in biological systems, researchers have been forced to develop new concepts and to rethink existing models of biological mechanisms. Tracking and understanding these changes in research practice and in biological theory will represent an important task for HPLS, not only to build a stronger response to the pandemic, but also to build a better understanding of the modern life sciences more generally.
Stephan Guttinger is a philosopher of biology based at LSE’s Centre for Philosophy of Natural and Social Science. Apart from looking at viruses and process ontology, he is also interested in how biologists create trustworthy knowledge, and how conceptual changes in the life sciences affect broader debates about science and health policy.
This is an edited version of Stephan’s recent paper. Covid-19 and the need for more history and philosophy of RNA. HPLS 43, 42 (2021).









































































































This is a VERY accurate post by Stephan Guttinger. Accord: https://blogdredd.blogspot.com/2022/02/quantum-biology-2.html
Great blog! Really enjoyed reading it. The content was informative and presented in such an engaging way!
Impressive content! This is a valuable resource. Thanks for sharing!